
This article explores integrating Large Language Models and Knowledge Graphs for advanced healthcare diagnostics, demonstrating future AI capabilities.
Introduction
Harnessing the power of AI, this article explores how Large Language Models (LLMs) like OpenAI’s GPT can analyze data from Knowledge Graphs to revolutionize data interpretation, particularly in healthcare. We’ll illustrate a use case where an LLM assesses patient symptoms from a Knowledge Graph to suggest diagnoses, showcasing LLM’s potential to support medical diagnostics with precision.
Brief Introduction Into Large Language Models (LLMs)
Large Language Models (LLMs), such as OpenAI’s GPT series, represent a significant advancement in the field of artificial intelligence. These models are trained on vast datasets of text, enabling them to understand and generate human-like language.
LLMs are adept at understanding complex questions and providing appropriate responses, akin to human analysis. This capability stems from their extensive training on diverse datasets, allowing them to interpret context and generate relevant text-based answers.
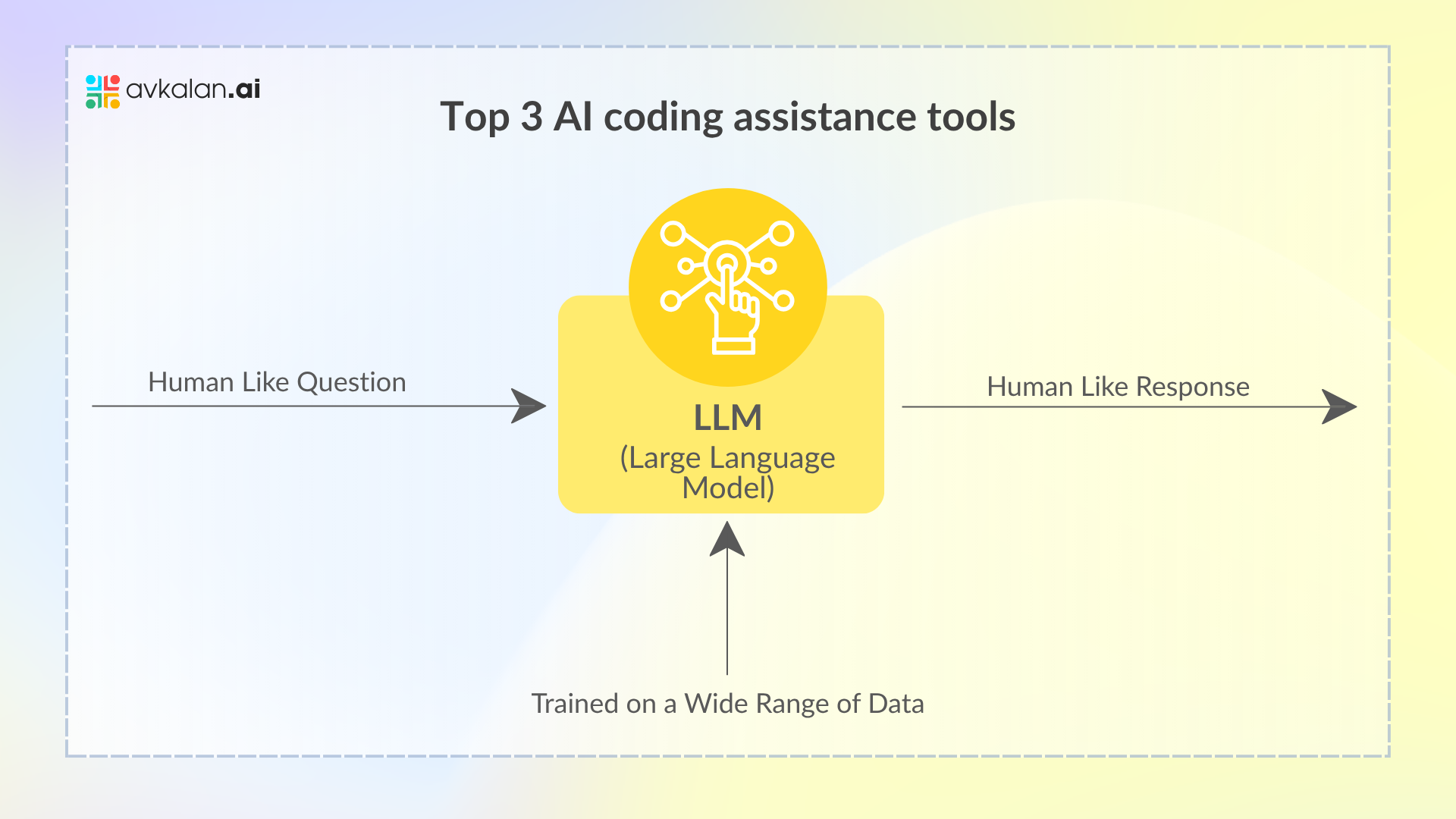
While LLMs possess advanced data processing capabilities, their effectiveness is often limited by the static nature of their training data. Knowledge Graphs step in to fill this gap, offering a dynamic and continuously updated source of information. This integration not only equips LLMs with the latest data, enhancing the accuracy and relevance of their output but also empowers them to solve more complex problems with a greater level of sophistication. As we harness this powerful combination, we pave the way for innovative solutions across various sectors that demand real-time intelligence, such as the ever-fluctuating stock market.
Exploring Knowledge Graphs and How LLMs Can Benefit From Them
Knowledge Graphs represent a pivotal advancement in organizing and utilizing data, especially in enhancing the capabilities of Large Language Models (LLMs).
Knowledge Graphs organize data in a graph format, where entities (like people, places, and things) are nodes, and the relationships between them are edges. This structure allows for a more nuanced representation of data and its interconnected nature.
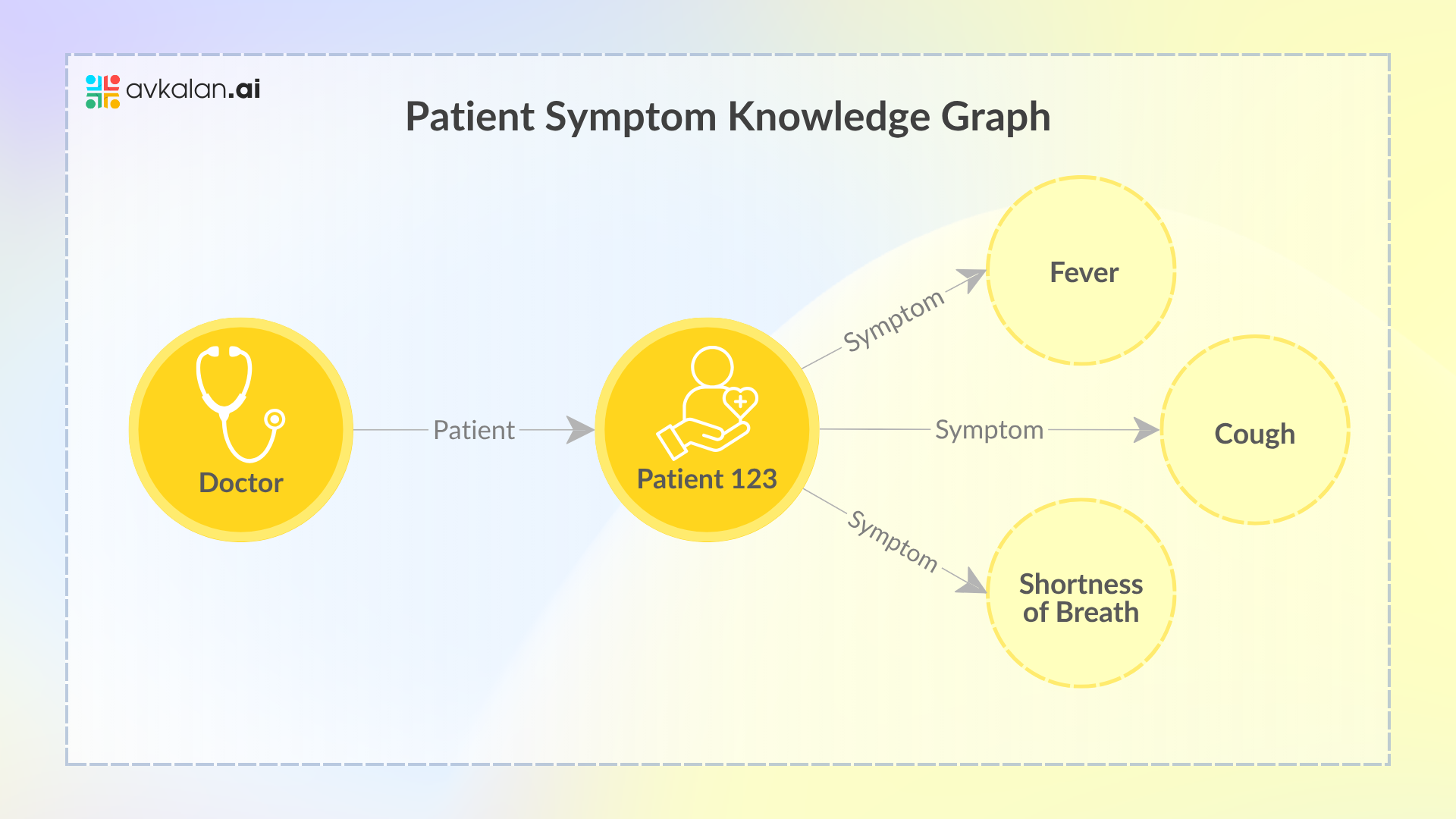
Take the above Knowledge Graph as an example.
- Doctor Node: This node represents the doctor. It is connected to the patient node with an edge labeled “Patient,” indicating the doctor-patient relationship.
- Patient Node (Patient123): This is the central node representing a specific patient, known as “Patient123.” It serves as a junction point connecting to various symptoms that the patient is experiencing.
- Symptom Nodes: There are three separate nodes representing individual symptoms that the patient has: “Fever,” “Cough,” and “Shortness of breath.” Each of these symptoms is connected to the patient node by edges labeled “Symptom,” indicating that these are the symptoms experienced by “Patient123.”
To simplify, the Knowledge Graph shows that “Patient123” is a patient of the “Doctor” and is experiencing three symptoms: fever, cough, and shortness of breath. This type of graph is useful in where it’s essential to model the relationships between patients, their healthcare providers, and their medical conditions or symptoms. It allows for easy querying of related data—for example, finding all symptoms associated with a particular patient or identifying all patients experiencing a certain symptom.
Practical Integration of LLMs and Knowledge Graphs
Step 1: Installing and Importing the Necessary Libraries
In this step, we’re going to bring in two essential libraries: rdflib for constructing our Knowledge Graph and openai for tapping into the capabilities of GPT, the Large Language Model.
| !pip install rdflib
!pip install openai==0.28 import rdflib import openai |
Step 2: Import your Personal OPENAI API KEY
This step is crucial for authenticating your requests to the OpenAI API. You must sign up with OpenAI and obtain an API key, which you’ll use in your code to make API calls.
| openai.api_key = “Insert Your Personal OpenAI API Key Here” |
Step 3: Creating a Knowledge Graph
Here, you initiate an empty RDF graph using rdflib. This graph will later be populated with data about patients and their symptoms, structured in a way that mirrors the relationships between entities (patients, symptoms) in the real world.
| # Create a new and empty Knowledge graph
g = rdflib.Graph() # Define a Namespace for health-related data namespace = rdflib.Namespace(“http://example.org/health/”) |
Step 4: Adding data to Our Graph
In this step, we will define a function to add data to the Knowledge Graph. This example specifically adds data for a patient, identified as “Patient123”, along with their symptoms. Each patient and symptom is represented as a node in the graph, and their relationship (a patient has a symptom) is represented as an edge.
| def add_patient_data(patient_id, symptoms):
patient_uri = rdflib.URIRef(patient_id)
for symptom in symptoms: symptom_predicate = namespace.hasSymptom g.add((patient_uri, symptom_predicate, rdflib.Literal(symptom))) # Example of adding patient data add_patient_data(“Patient123”, [“fever”, “cough”, “shortness of breath”]) |
Step 5: Identifying the get_stock_price function
In this step, we will define a function to query the Knowledge Graph for a patient’s symptoms using SPARQL, a query language for RDF. This function takes a patient ID, constructs a SPARQL query to fetch all symptoms associated with that patient, and returns the symptoms.
| def get_patient_symptoms(patient_id):
# Correctly reference the patient’s URI in the SPARQL query patient_uri = rdflib.URIRef(patient_id) sparql_query = f””” PREFIX ex: <http://example.org/health/> SELECT ?symptom WHERE {{ <{patient_uri}> ex:hasSymptom ?symptom. }} “”” query_result = g.query(sparql_query) symptoms = [str(row.symptom) for row in query_result] return symptoms |
Step 6: Identifying the generate_llm_response function
The generate_daignosis_response function takes as inputs the user’s name along with the list of symptoms extracted from the graph. Moving on, the LLM uses such data in order to give the patient the most appropraite daignosis.
| def generate_diagnosis_response(patient_id, symptoms):
symptoms_list = “, “.join(symptoms) prompt = f”A patient with the following symptoms – {symptoms_list} – has been observed. Based on these symptoms, what could be a potential diagnosis?”
# Placeholder for LLM response (use the actual OpenAI API) llm_response = openai.Completion.create( model=”text-davinci-003″, prompt=prompt, max_tokens=100 ) return llm_response.choices[0].text.strip() # Example usage patient_id = “Patient123” symptoms = get_patient_symptoms(patient_id) if symptoms: diagnosis = generate_diagnosis_response(patient_id, symptoms) print(diagnosis) else: print(f”No symptoms found for {patient_id}.”) |
Output: The potential diagnosis could be pneumonia. Pneumonia is a type of respiratory infection that causes symptoms including fever, cough, and shortness of breath. Other potential diagnoses should be considered as well and should be discussed with a medical professional.
As demonstrated, this function effectively uses the LLM to interpret the patient’s symptoms and suggest a diagnosis, showcasing the potential of combining structured knowledge (from the Knowledge Graph) with the generative and interpretative capabilities of LLMs.
This process highlights the bridge between structured data and natural language understanding, offering a glimpse into the future of AI-assisted diagnostics and decision-making.
Knowledge Graphs and Fusion Queries
Lastly, we will state how Knowledge graphs can be utilized with approaches such as fusion queries. Fusion queries refer to the process of simultaneously querying across different data sources, such as Knowledge Graphs, relational databases, and NoSQL databases, to obtain a unified result that incorporates insights from each source. This method allows for a more nuanced and comprehensive analysis than querying each source in isolation.
Fusion queries provide a more complete view by combining the rich, interconnected data of Knowledge Graphs with the vast storage and diverse data types available in other databases. Thus they enable applications to understand the context better and relationships within the data, leading to more accurate and relevant results.
For example, lets build on the healthcare scenario outlined in the article, where a Large Language Model (LLM) assesses patient symptoms from a Knowledge Graph to suggest diagnoses, let’s enhance this with a fusion query that incorporates additional data sources.
This approach will not only utilize the structured data from the Knowledge Graph but also integrate patient historical records from a relational database and unstructured clinical notes stored in a NoSQL database, thereby enriching the diagnosis process.
As in the original scenario, the Knowledge Graph is queried to identify patient “Patient123” and their symptoms. With the knowledge graph data being: Symptoms of “Patient123” – fever, cough, and shortness of breath.
Concurrently, the system queries a relational database for “Patient123″‘s medical history, focusing on past diagnoses, treatments, and outcomes. The relational database reveals “Patient123” has a history of asthma and allergies.
Finally, the gathered data – current symptoms from the Knowledge Graph, medical history from the relational database, and insights from clinical notes – are combined to form a comprehensive context for diagnosis.
Conclusion
In summary, the collaboration of Large Language Models and Knowledge Graphs presents a substantial advancement in the realm of data analysis. This article has provided a straightforward illustration of their potential when working in tandem, with LLMs efficiently extracting and interpreting data from Knowledge Graphs.
As we further develop and refine these technologies, we hold the promise of significantly improving analytical capabilities and informing more sophisticated decision-making in an increasingly data-driven world.
